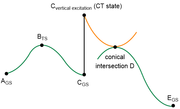Multiwfn forum
Multiwfn official website: //www.umsyar.com/multiwfn. Multiwfn forum in Chinese: http://bbs.keinsci.com/wfn
You are not logged in.
- Topics:Active|Unanswered
#1Re:Quantum Chemistry»Solvent participation after transition state»2026-02-02 02:04:03
Thank you for the answer.
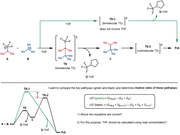
To explain my purpose more specifically, I attached another figure here, with a subsequent chemical step and an additional pathway.
Basically, my confusion is coming from the fact that the reactants and the final product are exactly same in the two pathways, but the different "timing" of the THF complexation (to BF3) in the one-step pathway and the two-step pathway.
Could you please further clarify the additional two questions that are included in the figure?
#2Quantum Chemistry»Solvent participation after transition state»2026-01-31 09:09:34
- wham09
- Replies: 3
-
Dear Prof. Lu,
I'd like to ask for your advice about how to calculate the free energy change of a solution-phase reaction when solvent coordination seems to occur after the transition state.
For convenience of understanding what I mean, I attached the reaction diagram, explanations, and questions as a figure.
My questions are all in the figure, but I will also paste them below.
4. What would be the correct way here?
4-1) THF coordination to D happens after this reaction. dG = (G_C + G_D) – (G_A + G_B)
4-2) THF coordination to D happens during product formation. dG = (G_C + G_D-THF) – (G_A + G_B + G_THF)
5. If 4-2 is correct, should G_THF be calculated using the concentration as 12.3 M (neat solvent molarity) rather than 1.0 M? (when plugging into the dG equation)
Thank you very much in advance.
#3Quantum Chemistry»Adjusting halogen radius for geometry optimization»2026-01-31 03:24:00
- wham09
- Replies: 1
-
Dear Prof. Lu,
I am calculating a toluene-solvated organometallic transition state that contain half-cleaved palladium-bromide bond. I was optimizing the geometry of this TS using PBE0-D3(BJ)/ma-SVP (PCM, toluene) level of theory, and observed oscillating behavior.
Out of curiosity, I modified the PCM radius of bromine atom to 2.60 by using modifysph command, which is what was done in Truhlar's SMD18 paper (https://doi.org/10.1002/chem.201803652).
Of course, what Truhlar did was optimizing the geometries of his compounds using M06-2X/def2-TZVP (def2-TZVPD for bromine). I knew this, but I still chose to optimize my compound using PBE0-D3(BJ)/ma-SVP (PCM, toluene) and only change the PCM radius of bromine.
By doing this, the TS optimization was successful without oscillating behavior. Do you think that this could actually be a generally better way to calculate bromine-containing molecules, even when using functionals and basis sets that are different from the original SMD18 paper?
#4Re:Multiwfn and wavefunction analysis»w-tuning for n-th excited state»2026-01-30 00:33:07
Thank you for the answer.
I’d like to ask just one follow-up question.
The ground state of my molecule is doublet D0, and the first excited state (D1) is characterized by alpha-HOMO (30A) to alpha-LUMO (31A), while beta-HOMO stays 29B.
In this case, when I calculate N, N+1, and N-1 states for w-tuning, should I set the N+1 state as triplet? In this way the N+1 state will have the 31A populated.
#5Multiwfn and wavefunction analysis»w-tuning for n-th excited state»2026-01-28 02:42:36
- wham09
- Replies: 3
-
Dear Prof. Lu,
I am currently trying to conduct electron excitation analysis for the 5th excited state of my molecule.
The 5th excitation is characterized by charge transfer from alpha-HOMO to alpha-LUMO+3.
If I wanted to do w-tuning of long-range-corrected functional for this state, should I then look at N and N-1 (for HOMO), and separately, N+7 and N+6 (for LUMO+3)?
#6Re:Quantum Chemistry»Questions on how to rigorously measure free energies in solution»2026-01-20 05:49:12
I would be really grateful if you could explain the answer to Q1 a little more. The way I understand is that the G in solvent phase is as below:
G_sol = G_gas + dG_solv + 1.89
Because G_gas = E_gas + dG_corr_gas,
G_sol = (E_gas + dG_corr_gas) + dG_solv + 1.89
Here, dG_corr_gas should be the correction for gas-phase G. Then, shouldn’t I actually put E_gas into Shermo to obtain G_gas first, and then manually add dG_solv and 1.89kcal/mol?
#7Quantum Chemistry»Questions on how to rigorously measure free energies in solution»2026-01-20 03:23:59
- wham09
- Replies: 3
-
Dear Prof. Lu,
I did the following calculations for a ground-state molecule:
calc1. Geometry optimization (PCM model) + frequency calculation (B3LYP-D3(BJ) / def2-SVP, PCM)
calc2. Single point calculation (gas-phase) using calc1 geometry (B3LYP-D3(BJ) / def2-TZVP)
calc3. Single point calculation (gas-phase) using calc1 geometry (M05-2X / 6-31G*)
calc4. Single point calculation (SMD model) using calc1 geometry (M05-2X / 6-31G*, SMD)
From calc2, I obtained E_gas.
From calc3 and calc4, I obtained dG_solv.
If I understood correctly, the free energy is calculated as:
G = G_gas + dG_solv + 1.89
= (E_gas + dG_corr_gas) + dG_solv + 1.89
So I utilized Shermo on calc1 output for ZPVE scaling and msRRHO treatment, and obtained dG_corr_gas.
But here are my questions:
1. Am I correct to put E_gas (from calc2) into the Shermo's E value in the setting.ini file, rather than E_gas + dG_solv?
2. For calc3 and calc4 (using M05-2X functional), should superfine grids be used, as they sometimes should be for M06-2X?
3. In line with Q3, if I choose to do calc3 and calc4 using M06-2X instead, should I use superfine grids consistently?
4. calc1 was done with PCM because the geometry was sensitive to solvation. But then, is the Shermo result using the calc1 output truly the dG_corr_gas?
5. As an alternative, I did another frequency calculation (calc5) in the gas-phase using the calc1 geometry, but when I did this, I observed imaginary frequencies (expectedly). Would using Shermo on the calc5 output be more appropriate than using the calc1 output anyways?
6. Is there a truly ideal solution to Q4 and Q5? I don't care if it is cumbersome, I just want to know.
Thank you very much again for your time.
#8Re:Multiwfn and wavefunction analysis»CT excited state energy and wavefunction analysis»2026-01-15 05:22:59
About question 1: This is actually what I am most confused, because I thought I was correct based on your blog posts. Could you please correct me if I’m interpreting your texts in a wrong manner? (or wrong translation?) See below:
In your blog post “On the diffuse function and the "month" basis set (//www.umsyar.com/119)”,
1. When is a diffusion function needed?
…
• Adding dispersion is very important and strongly recommended:
Calculate the weak interaction energy (if using a 4-zeta level basis set and not involving anions, then dispersion is not necessary)
…
• Adding dispersion can improve results, and should be done
optimize the structure of anionic or weakly interacting systems;
...
Also in “On the choice of basis sets in quantum chemistry (//www.umsyar.com/336)”,
2. Selection of different task basis sets
2.2 Weak Interactions
…
The greater the proportion of dispersive interactions, the more necessary a diffuse function is required, and the more important the BSSE problem needs to be considered.
Basis set recommendations for weak interaction energy calculations:
Minimum: 6-31+G** or ma-def 2-SVP for DFT
…
Many thanks in advance!
#9Re:Multiwfn and wavefunction analysis»CT excited state energy and wavefunction analysis»2026-01-14 04:37:42
I’d be really grateful if you could provide some additional feedbacks, basically whether you agree to the following statements.
1. The structure (of which I’m studying the CT state) is a bimolecular interacting complex, which features intermolecular pi-stacking between two aromatic groups, and CT occurs between these two rings. In this case, even without significantly negatively charged atoms, it is recommended to add diffuse functions.
2. In the case of 1. or in case there are actually highly negatively charged atoms, I just have to accept that the method I have to use to get accurate energy (using diffuse functions) is not going to permit analysis of correct LUMO or LUMO+1.
Thank you very much.
#10Multiwfn and wavefunction analysis»visualizing exchange and correlation holes in sobEDA framework»2026-01-13 19:07:07
- wham09
- Replies: 1
-
Dear Prof. Lu,
I am wondering if I could somehow monitor the electron density change over the course of sobEDA steps so that I could separately examine exchange and correlation effects. Changes beginning from the promolecular state should be trivial, I would just have to substract e-density function between promol/frozen and frozen/final.
But ideally, I also want to dissect the first step (fragments to promolecular) into multiple different electronic structures to separate electrostatic, exchange, correlation, and dispersion effects on the electron density. What I could think of is repeating the fragment-to-promolecular step 1) with HF level, and 2) with the original level but without DFT-D3. (Original level I used for the whole sobEDA was PBE0-D3(BJ)/ma-TZVP).
Please let me know if this whole thing is just a stupid idea, or if it might be possible in a more sophisticated manner. Thank you very much!
#11Re:Multiwfn and wavefunction analysis»CT excited state energy and wavefunction analysis»2026-01-09 02:36:36
Thank you for the reply,
If you don’t mind, could you help me understand this in a little more detail? Because I thought that in a charge-transfer state where certain atoms (that received charge) are significantly negatively charged, at least those atoms should need to be treated with diffuse functions.
#12Multiwfn and wavefunction analysis»CT excited state energy and wavefunction analysis»2026-01-08 03:11:33
- wham09
- Replies: 7
-
Dear Prof Lu,
I have a neutral doublet structure (D0) whose first excited state (D1) turns out to be a charge-transfer state, which shows charge-separation (significantly negative charges on specific atoms and positive charges on other atoms). From what I learned, I would need to add diffuse functions to accurately measure the excitation energy.
At the same time, I want to study the D1 state's electronic state, examine molecular orbitals, etc. But I noticed that with diffuse functions used, the alpha-HOMO in the D1 state (alpha-LUMO in the D0 state) is severely broken in shape. Only when I do not add diffuse functions I see excitation to the correctly shaped orbital.
How should I resolve both energy evaluation and accurate wavefunction analysis at the same time?
#13Re:Multiwfn and wavefunction analysis»Miscellaneous questions on TDDFT/electron-hole analysis»2025-11-09 08:46:46
question 8: From what I understood your comments, in the figure that I attached (green: non-excited PES, orange: excited PES), is it correct that calculating accurate relative Gibbs free energies of all 6 points under the same calculation level is not feasible?
Especially because, for my specific molecule, the CT state of interest is assigned as the 2nd excited state when calculated with wB97XD, but is correctly (I think) assigned as the 1st excited state only when the w-tuned LC-wPBE is used.
question 9: Also related to question 8, if I optimize the MECI structure with CASSCF, is it possible to calculate its delta G relative to other states on the reaction path calculated by DFT methods?
Additional question about Q9: As I explained above, the CT state of my molecule is assigned differently depending on the functional. So I'm not confident that CASSCF will assign the CT state as the 1st excited state. Then, should I increase the number of electrons and orbitals in the active space to be safe?
Additional question (new): I would think that state-specific solvation of the vertical excitation state will affect vibrational frequencies differently (in principle) from the linear response. Can I do excited state freq calculation under this exact state-specific environment? Perhaps do TD=read from the state-specific solvated TD calculation chk?
#14Re:Multiwfn and wavefunction analysis»Miscellaneous questions on TDDFT/electron-hole analysis»2025-11-08 13:03:43
About question 1: To be more specific, after TD optimization is complete, and then if I want to run single-point calculation of the excited state to obtain total Gibbs free energy (following the procedure you suggested for the question 7), would you recommend DFT-D3(BJ) for the functionals I mentioned?
About question 5: Just like question 1, for single-point calculation of the optimized excited state, SMD is preferred, right?
About question 8: I guess the question was not specifically about CAM-B3LYP. For example, I could use LC-wPBE with w-tuning specific to the structure of B*. In this case, I thought that the specifically tuned LC-wPBE is probably not adequate for calculation of other ground states and transition states in the whole reaction path (which could be 10~20 steps), compared to more general functionals such as PBE0-D3(BJ) or B3LYP-D3(BJ). Is it okay to use a more general functional for non-excited states and compare their free energies with the free energy of B* calculated by tuned LC-wPBE?
About question 9: I do have knowledge on conical intersections, although not an expert. My guess is perform relaxed scans (from the structure of B* to C) with 1) standard and 2) TD settings, and find out a scan point where the two energies are most similar? Could you let me know if there is a better, or a more appropriate, procedure?
#15Multiwfn and wavefunction analysis»Miscellaneous questions on TDDFT/electron-hole analysis»2025-11-07 03:48:11
- wham09
- Replies: 5
-
Dear Prof. Lu,
I hope to get help with several questions on TDDFT calculation for electron-hole analysis.
1. For TDDFT, whether the functional is PBE0, LC-wPBE, CAM-B3LYP, or wB97XD, would you generally recommend DFT-D3(BJ)?
2. Is LC-wHPBE really superior to LC-wPBE as Gaussian advertises?
3. Can the same w-tuning procedure be applied whether the ground-state is singlet, doublet, or triplet, and whether it is neutral, anionic, or cationic?
4. Can electron excitation and a bond-formation or bond-cleavage reaction be concerted? Can it be calculated?
5. Is it more recommended (or not recommended) to do TD calculations with SMD rather than PCM?
6. For excited state optimization, can I just use nosymm keyword and skip the manual arbitrary breaking of symmetry?
7. For calculation of total Gibbs energy of an optimized excited state, please check if what I describe below seems correct:
1) ES optimization (with GS checkpoint file available):
wB97XD/def2-SVP TD(Nstates=5, Root=2) Opt SCRF=solvent=dimethylsulfoxide geom=modify ... geometry modification
2) Take the structure and .chk file from 1). Run freq calculation:
wB97XD/def2-SVP TD(read, Nstates=5, Root=2) Freq SCRF=solvent=dimethylsulfoxide guess=read
3) Take the structure and .chk file from 1). Run SP calculation (gas-phase):
wB97XD/def2-TZVP TD(Nstates=5, Root=2) guess=read
4) Take the structure and .chk file from 1). Run SP calculation (gas-phase):
M052X/6-31G* TD(Nstates=5, Root=2) guess=read
5) Take the structure and .chk file from 1). Run SP calculation (solution-phase):
M052X/6-31G* TD(Nstates=5, Root=2) SCRF=solvent=dimethylsulfoxide guess=read
6) Calculate E = E(step 3) + E(step 5) - E(step 4). Put it into Shermo and use .out file from step 2. Get thermal correction.
8. Suppose I have a reaction pathway A -> TS-1 -> B -> B* -> TS-2 -> C. The excited state B* is a charge-transfer state, for which I would want to use a long-range-corrected functional, such as CAM-B3LYP. But CAM-B3LYP would be less appropriate for calculation of other states A, B, TS-1, and TS-2. But then, for reaction coordinate energy calculations, shouldn't the functional be consistently used for every molecule from A to C?
9. For the TS-2 between B* and C in the question 8, am I looking for "excited state of transition state"? I don't know how to think about it.
#16Multiwfn and wavefunction analysis»Electron excitation analysis of radicals»2025-11-06 09:24:51
- wham09
- Replies: 1
-
Dear Prof. Lu,
I am trying to study excitation of a doublet radical. There are some problems in using Multiwfn to analyze this problem, such as:
1) In the summary of excited states, all the [Multi.]'s are shown as "?" like below:
State: 1 Exc. Energy: 2.852 eV Multi.: ? MO pairs: 42461
2) Atom contribution to hole and electron results looks like this:
1(C ) Hole: 10.10 % Electron: NaN % Overlap: 0.00 % Diff.: NaN %
3) IFCT (Hirshfeld) results are strange. For example, the results for S2 state seem fine, but the results for S1 state are full with "NaN"s.
The input lines I used for TDDFT are as below:
%chk=[checkpoint from ground-state opt calculation using PBE0-D3(BJ)/ma-SVP]
#p scf=(maxcycle=300) scrf=(solvent=dimethylsulfoxide) uwB97XD/gen TD(nstates=5) iop(9/40=4) guess=read nosymm
[Title]
0 2
[optimized geometry]
[ma-TZVP basis functions]
I'd be grateful if you could tell what the problem is.
#17Re:Multiwfn and wavefunction analysis»Clarifications on diffuse functions for multiwfn»2025-11-05 08:14:13
About the additional question, sorry about the confusion. What I meant is as below:
My molecule is an ion-pair complex, consisting of a cation and an anion close together (total charge=0). The inter-fragment interaction between the cationic and the anionic counterparts is the primary interest. At the same time, the cationic and anionic charges in the two fragments are delocalized enough so that none of the individual atoms (in the structure of the ion-pair complex) are significantly negatively charged (ADCH charges of all atoms more positive than -0.5).
#18Re:Multiwfn and wavefunction analysis»Clarifications on diffuse functions for multiwfn»2025-11-03 07:37:03
Thank you very much for the clarification. I have a follow-up question for just #4:
My situation is as following:
1) The ion-pairing is important in my molecule. I want to study the interaction between the cationic and anionic counterparts.
2) One atom (out of total 70~80 atoms) is significantly negatively charged (calc. level: PBE0-D3(BJ) / ma-TZVP or def2-TZVP)
In the multiwfn manual on ETS-NOCV, it says “If there is no special reason, using diffuse functions is deprecated.” Does my situation fit into the “special reason”? Is ETS-NOCV absolutely meaningless when ma-TZVP is used?
Additional question: What if the ion-pairing is important, but there is no atom that is significantly negatively charged?
#19Multiwfn and wavefunction analysis»Clarifications on diffuse functions for multiwfn»2025-10-30 01:39:13
- wham09
- Replies: 5
-
Dear Prof. Lu,
I’d be really grateful if I could get additional clarifications on dealing with diffuse functions (and some additional questions on ZPE correction)
1. If I use the complete set of ma-def2 series that you provided, how should I cite it when I publish?
2. If the net charge of my molecule is overall -1, but each atom is not significantly negatively charged (ADCH charge > -0.5), diffuse functions are not required. Correct?
3. In my reaction A + B -> [A-B], some atoms in the product [A-B] are significantly negatively charged, but not in the reactant forms (A and B). For the sake of reaction energy calculation, should these atoms be calculated using diffuse functions for both reactant (A and B) and product ([A-B]) calculations?
4. A molecule is overall neutral, and there is no atom with a significant negative charge. But it is a tight ion-pair, in which the cation and anion counterparts are electrostatically bound and deform each other's structure. The binding is strong, but it is still non-covalent. Should diffuse functions be widely used?
5. When using Shermo for a solute, the sophisticated way of input for Shermo setting is:
E = E(gas, high-level basis set) + [E(SMD, M05-2X/6-31G*) - E(gas, M05-2X/6-31G*)] + 1.89 kcal/mol.
Am I correct?
6. When I do opt-freq calculations using SCRF(IEFPCM), the ZPE values are, not surprisingly, different from the values from gas-phase optimization. This will be same for the molecules in F38/10 database used to obtain the scale factor. Then, shouldn't the ZPE scale factor become different?
7. For wavefunction analysis of a solute, it is recommended to calculate with high-level basis set & SMD. But at the same time, for the calculation of the solute's solvation energy, the same structure should be calculated with M05-2X/6-31G*. Am I correct?
#20Re:Multiwfn and wavefunction analysis»Definition of “compatibility” with diffuse functions»2025-10-28 04:54:48
1. What is the bar for significantly negatively charged atom? mulliken or nbo charge? For example, the atom with the most negative nbo charge in my molecule has nbo charge of -0.59.
2. Would it work to use def2-TZVPD for only the significantly negatively charged atom and def2-TZVP for the rest?
3. If the molecule is overall neutral, but contains an atom that is significantly negatively charged, should diffuse functions be used?
4. If I use def2-TZVPD for only one atom in a molecule with total 14 atoms, would the output be incompatible with wavefunction analysis?
#21Re:Multiwfn and wavefunction analysis»Definition of “compatibility” with diffuse functions»2025-10-28 04:23:38
Would you also agree with “it retains the merit of diffuse basis sets for calculation of, for example, anions”?
#22Re:Multiwfn and wavefunction analysis»Definition of “compatibility” with diffuse functions»2025-10-28 04:00:47
I see. Then, would it be correct to state that def2-TZVP can be used for Multiwfn functions that are incompatible with diffuse functions, but it retains the merit of diffuse basis sets for calculation of, for example, anions?
#23Multiwfn and wavefunction analysis»Definition of “compatibility” with diffuse functions»2025-10-28 02:19:10
- wham09
- Replies: 7
-
Dear Prof. Lu,
I know that many functions of Multiwfn are incompatible with diffuse functions. But I’m confused with the definition of diffuse functions.
What I attached here is the basis set information for the same atom (boron) in the same molecule, left one using 6-311+g** and right one using def2-TZVP.
Although 6-311+g** is said to have diffuse functions (and def2-TZVP does not), the def2-TXVP setting also contains a S-type function with an exponent smaller than 0.1 (the last S-type in the right figure, exponent=0.06056). Also, the def2-TZVP setting contains an additional D-type function (exponent=0.199) that is more “diffuse” than the sole D-type function in the 6-311+g** setting (exponent=0.401).
So, what is the exact standard for calling a basis set diffuse or not?
#24Re:Quantum Chemistry»Frequency correction»2025-10-26 21:52:54
wham09 wrote:Dear Prof. Lu,
The Shermo code utilizes frequency scale factors for ZPE, U(T)-U(0), S, and CV. I can find the scale factors for ZPE in the literature, but the other three factors are rarely documented (especially, I could not find the factors for CV). So I'd like to ask if the following routine would work.
1. Fit the fundamental frequency factor for my functional/basis set using a database (F38/10 for example).
2. Run a opt-freq calculation for my molecule, and tabulate the computed harmonic frequencies.
3. Multiply the computed frequencies by the fitted fundamental frequency factor.
4. Put the results as the custom frequencies into a new freq calculation input with freq=(ReadFC, ReadIsotopes) keyword. (Or use scale keyword)
5. Use the output of the new freq calculation as the input file for Shermo, with the following settings:
5-1. sclZPE, sclheat, sclS, sclCV = 1.0
5-2. ilowfreq = 2 or 3Only ZPE scale factor is relatively important, you can simply set other factors to 1.
Thank you for the reply. As I said, I do understand that, but I just wanted to know if what I described would do the same thing as putting all scl values into Shermo?
#25Re:Multiwfn and wavefunction analysis»Help with understanding results of Shubin Liu's EDA»2025-10-25 16:36:31
Thank you for the clarification.
1. For sobEDAw, is it okay to use SDD (in combination with 6-31+G** or 6-311+G**)? I couldn't find any sobEDAw example in your paper and tutorial that deals with transition metals.
2. For both sobEDA and sobEDAw (actually, for any calculation), if I decided to use def2- family, would you recommend using a mixed basis set with SDD for TM-complex? Or use the def2- basis set solely?
3. If I want to do sobEDA or sobEDAw analysis using TPSSh/def2-TZVP, should I do the initial geometry optimization also using TPSSh/def2-family? I guess this question is also relevant to any calculation.
#26Re:Quantum Chemistry»Frequency correction»2025-10-25 09:45:27
Additional questions:
1. I understand that unlike sclZPE, other scale factors are normally close to unity. But I still want to understand how they're derived. From what I read in Moran/Radom paper (doi/10.1021/jp073974n), sclheat and sclS cannot be obtained by simply fitting U_calc and U_exp (or S_calc and S_exp) linearly. Am I correct?
2. Can I get any reference where the scale factors for CV are derived? I could never find it.
3. I tried fitting the frequency scale factor myself. For some of the molecules in F38/10 set, the degenerate frequencies appeared to be different from each other. For example, the frequencies corresponding to the 3rd mode (pi_u symmetry) of CO2 are 646.6657 and 646.6429 in my calculation output. Did I do something wrong? Or should I just average them?
4. If what I want at the end is G(sol), should I put into Shermo the E value from SCRF calculation, rather than gas-phase single-point?
#27Quantum Chemistry»Frequency correction»2025-10-25 02:32:37
- wham09
- Replies: 4
-
Dear Prof. Lu,
The Shermo code utilizes frequency scale factors for ZPE, U(T)-U(0), S, and CV. I can find the scale factors for ZPE in the literature, but the other three factors are rarely documented (especially, I could not find the factors for CV). So I'd like to ask if the following routine would work.
1. Fit the fundamental frequency factor for my functional/basis set using a database (F38/10 for example).
2. Run a opt-freq calculation for my molecule, and tabulate the computed harmonic frequencies.
3. Multiply the computed frequencies by the fitted fundamental frequency factor.
4. Put the results as the custom frequencies into a new freq calculation input with freq=(ReadFC, ReadIsotopes) keyword. (Or use scale keyword)
5. Use the output of the new freq calculation as the input file for Shermo, with the following settings:
5-1. sclZPE, sclheat, sclS, sclCV = 1.0
5-2. ilowfreq = 2 or 3
#28Re:Multiwfn and wavefunction analysis»Help with understanding results of Shubin Liu's EDA»2025-10-23 03:49:36
I'm a little confused because in the 5 template.gjf files,
The templates for b3lyp, blyp, TPSSTPSS: Do not contain the iop keyword.
The template for BHandHLYP contains: IOp(3/174=1000000,3/175=1035400,3/177=279300,3/178=4961500)
The template for TPSSh contains: IOp(3/174=1000000,3/175=2238200,3/177=452900,3/178=4655000)
So, are these options specific to each functional? or a combination of functional/basis set?
#29Re:Multiwfn and wavefunction analysis»Help with understanding results of Shubin Liu's EDA»2025-10-22 07:53:36
Just an additional technical question about sobEDA:
I noticed that among the sobEDA tutorials, the template gjf files for metal-containing systems (chromium alkylidene and copper/H2O) contain an additional keyword IOp(3/174=1000000,3/175=2238200,3/177=452900,3/178=4655000), which I believe is additional dispersion adjustment. Should this keyword (with these exact numbers) be applied to any organometallic species?
#30Re:Multiwfn and wavefunction analysis»Help with understanding results of Shubin Liu's EDA»2025-10-21 10:50:47
1. I looked into the sobEDA script file and concluded that the best option for me is using Multiwfn only to combine wavefunctions of fragments and generate promolecular & frozen input files, then run g16 jobs, and then tabulate the results in the l608 section myself. Could you confirm that this will work?
2. I now understand what you mean. Thank you very much!